Recipients: Joff Silberg, Vicky Yao
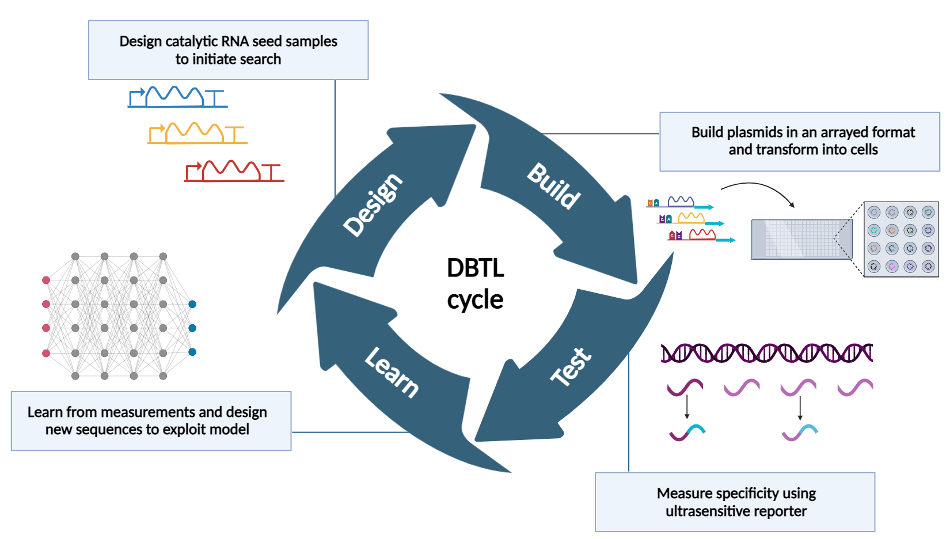
The overuse and non-specific nature of chemical antibiotics has led to an explosion of drug-resistant pathogens, limiting the effectiveness of this class of therapeutics. Genetically-encoded antibiotics represent an emerging strategy to address this challenge. These antibiotics can use unique genetic signatures in microbial communities to switch on the production of a toxic protein. By using genetic signatures specific to pathogens as triggers for toxic protein production, genetically-encoded antibiotics have the potential to selectively induce cell death, killing the pathogen while leaving the beneficial bacteria in the community unharmed. However, the design space for genetically-encoded antibiotics is massive, and it remains challenging to identify optimal designs to ensure precise pathogen targeting with minimal off-target effects. To overcome this, the Silberg and Yao labs are collaborating to incorporate machine learning and language models to guide and enhance the design-build-test-learn cycle. This integration aims to accelerate the identification and optimization of effective antibiotic designs. More specifically, the combinatorial libraries of antibiotics are being designed and selected for functional variants, while computational models are used to prioritize targets and refine the selection process. This combined approach is expected to uncover the principles governing optimal target selection, leading to the rapid development of next-generation genetically-encoded antibiotics. By focusing on the precise targeting of pathogens, these antibiotics hold the potential to revolutionize the treatment of infectious diseases, reduce the emergence of drug-resistant strains, and significantly improve global health outcomes.