Recipients: Jeffrey J. Tabor, Han Xiao, Elizabeth Gardner
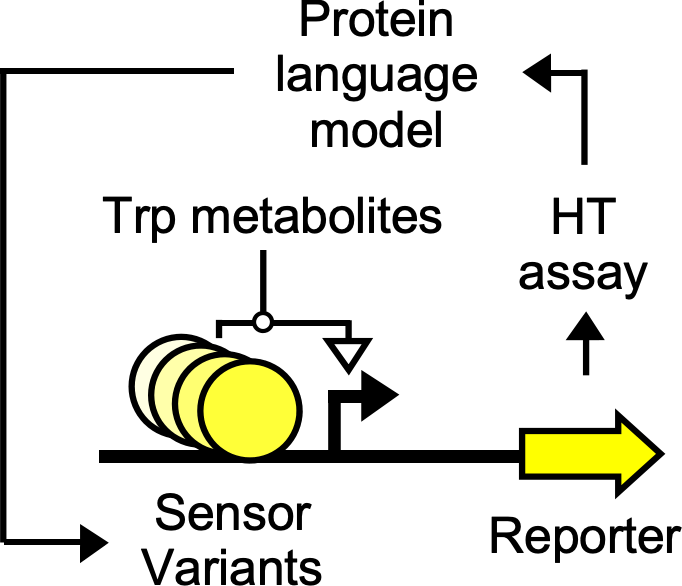
Metabolic pathways encoded in the human genome convert the amino acid tryptophan into downstream metabolites that are linked to immune and nervous system function, cancers, and cardiovascular disease among other processes. Our long-term goal is to engineer genetically-encoded sensors that enable living cells to specifically sense tryptophan pathway metabolites in order to study their physiological roles in situ and detect and treat diseases to which these molecules are linked. In this project, we will leverage a new high-throughput protein engineering method to redesign the ligand binding pocket of a well characterized transcription factor protein that senses a specific tryptophan metabolite to sense other pathway metabolites in a highly specific manner. Initially, we will introduce mutations that we have rationally chosen based on the known structure of the transcription factor:ligand complex and screen the resulting mutants against the new metabolites. Then, we will use a high-throughput gene expression assay to characterize the response of these mutants to the new metabolites, and feed the resulting data into a protein language model which will guide the design of subsequent rounds of mutations. We will iterate upon this process until we construct a panel of sensor mutants with novel specificities toward tryptophan pathway metabolites. The sensors we construct can be introduced into bacteria or mammalian cells in future studies and used to quantify the levels of these metabolites in different tissues in vivo. They can then be linked to reporter genes to enable fundamental studies and therapeutic genes to ameliorate disease states.